ICLAC Register of Misidentified Cell Lines
This resource is curated by ICLAC and lists cell lines that are known to be cross-contaminated or otherwise misidentified. The ICLAC Register of Misidentified Cell Lines has its own page with more information here.
Online Databases and Search Tools for Cell Line STR Profiles
These resources consist of databases containing cell line STR profiles and search tools that allow the user to enter one or more STR profiles for similarity-based searches. For more information, see the links and references that are listed below.
The Cellosaurus is a knowledge resource on cell lines. It attempts to describe all cell lines used in biomedical research. For each cell line (currently ~120,000) the Cellosaurus provides a wealth of information.
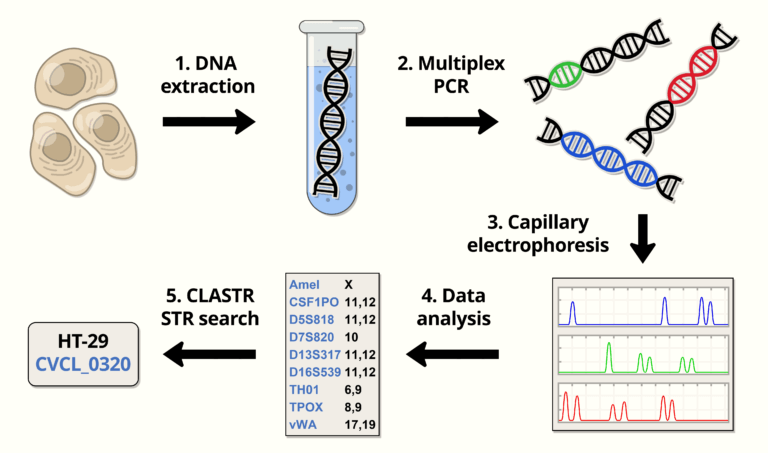
CLASTR is a STR similarity search tool that allows to compare one or more STR profiles from human, dog or mouse cell lines with those available in the Cellosaurus (~7000 profiles).
AuthentiCell allows you to search for similarities between your cell line samples and an extensive number of human cell line STR profiles stored in the database. AuthentiCell is a service provided by the European Collection of Authenticated Cell Cultures (ECACC).
Leibniz Institute DSMZ operates a user-friendly interface to query its database of STR profiles, brought together in collaboration with cell banks ATCC, JCRB, and RIKEN as a human cell line cross-contamination initiative.
The Cell Line Integrated Molecular Authentication (CLIMA) database is the result of an integration effort aimed at including all certified STR profiles of human cell lines in a unique database. Its main feature is cell line identification according to the standard ANSI/ATCC ASN-0002-2011, but it also provides features for searching profiles by cell line name and by locus.
As part of their continuing efforts to characterize and authenticate the cell lines in the Cell Biology collection, ATCC has developed a comprehensive database of short tandem repeat (STR) DNA profiles for all of their human cell lines.
The NCBI BioSample database archives STR profiles and descriptions for several thousand cell lines, as provided by cell line repositories. The database is administered by the US National Library of Medicine, National Institutes of Health.
Publications that describe these databases and search tools include:
Bairoch A (2018) The Cellosaurus, a cell-line knowledge resource. J Biomol Tech 29: 25-38. PMID: 29805321.
Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, Yaschenko E, Ostell J (2012) BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res 40: D57-63. PMID: 22139929.
Dirks WG, MacLeod RA, Nakamura Y, Kohara A, Reid Y, Milch H, Drexler HG, Mizusawa H (2010) Cell line cross-contamination initiative: an interactive reference database of STR profiles covering common cancer cell lines. Int J Cancer 126: 303-4. PMID: 19859913.
Robin T, Capes-Davis A, Bairoch A (2019) CLASTR: the Cellosaurus STR similarity search tool, a precious help for cell line authentication. Int J Cancer, Epub 24 August. PMID: 31444973.
Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B (2009) Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res 37(Database issue): D925-D932. PMID: 18927105.
Somaschini A, Amboldi N, Nuzzo A, Scacheri E, Ukmar G, Ballinari D, Malyszko J, Raddrizzani L, Landonio A, Gasparri F, Galvani A, Isacchi A, Bosotti R (2013) Cell Line Identity Finding by Fingerprinting, an Optimized Resource for Short Tandem Repeat Profile Authentication. Genet Test Mol Biomarkers 17: 254-9. PMID: 23356232.
Publications or Websites with Cell Line STR and SNP Data
Publications and websites that contain STR profiles and/or SNP data from cell line panels include:
COSMIC Cell Lines Project – QC spreadsheet
http://cancer.sanger.ac.uk/cancergenome/projects/cell_lines/about
Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD (2010) Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst 102: 1310-21. PMID: 20679594.
Liang-Chu MM, Yu M, Haverty PM, Koeman J, Ziegle J, Lee M, Bourgon R, Neve RM (2015) Human Biosample Authentication Using the High-Throughput, Cost-Effective SNPtraceTM System. PLoS One 10: e0116218. PMID: 25714623.
Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, Weinstein JN (2009) DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther 8: 713-24. PMID: 19372543.
Yu M, Selvaraj SK, Liang-Chu MMY, Aghajani S, Busse M, Yuan J, Lee G, Peale F, Klijn C, Bourgon R, Kaminker JS, Neve RM (2015) A resource for cell line authentication, annotation and quality control. Nature 520: 307-11. PMID: 25877200.
Publications or Websites with Cell Line Names
Publications and websites with information on cell line names include:
Cell Line Ontology (CLO)
http://www.clo-ontology.org/
Human Pluripotent Stem Cell Registry (hPSCreg)
http://hpscreg.eu/
HyperCLDB
http://bioinformatics.hsanmartino.it/hypercldb/
Kurtz A, Seltmann S, Bairoch A, Bittner MS, Bruce K, Capes-Davis A, Clarke L, Crook JM, Daheron L, Dewender J, Faulconbridge A, Fujibuchi W, Gutteridge A, Hei DJ, Kim YO, Kim JH, Kokocinski AK, Lekschas F, Lomax GP, Loring JF, Ludwig T, Mah N, Matsui T, Müller R, Parkinson H, Sheldon M, Smith K, Stachelscheid H, Stacey G, Streeter I, Veiga A, Xu RH (2018) A standard nomenclature for referencing and authentication of pluripotent stem cells. Stem Cell Reports 10: 1-6. PMID: 29320760.
Yu M, Selvaraj SK, Liang-Chu MMY, Aghajani S, Busse M, Yuan J, Lee G, Peale F, Klijn C, Bourgon R, Kaminker JS, Neve RM (2015) A resource for cell line authentication, annotation and quality control. Nature 520: 307-11. PMID: 25877200.