Author | Erin Hall, ICLAC member
This case study will delve into the history and identity of Chang Liver. We hope it will serve as a notice to researchers who continue to use the cell line incorrectly as an in vitro model for normal liver.
Chang liver can be considered “notorious” because the contaminating cell line (the famous HeLa) is well known to the community, and because the misidentification of Chang liver has been known for more than half a century.
This case study is available for download | Chang Liver Case Study PDF
History of Chang Liver
Dr R. Shihman Chang is considered a cell culture pioneer. With his work beginning in the 1950’s; he was part of the first wave of researchers who established immortalized lymphoid cell lines through the use of Epstein Barr Virus (EBV, Ref. 1). He was particularly interested in creating cell lines that supported the growth of viruses in order to study them in vitro.
The cell line that has come to be known as “Chang Liver” originated from the biopsy of a patient during an exploratory laparotomy in the early 1950s (2). Information on the patient from which Chang Liver was derived is scarce (3):
Since the goal was merely to obtain a sufficient number of cells that
would support the growth of certain viruses under study, no effort was made to record the sex, race, age, and medical diagnosis of the tissue donor.”
– Dr Chang-
The “Chang Liver” cell line was deposited at the American Type Culture Collection in December 1962 (ATCC® catalogue #CCL-13™). Chang Liver was deposited for distribution to other scientists, who for their own studies, were in need of an immortalized “normal liver” in vitro model, a very rare commodity during these early years as cell culture techniques were being devised and implemented.
Chang Liver: HeLa Cells?
In 1967, Stanley Gartler, a researcher at the University of Washington, presented evidence that Chang Liver (among many other supposedly “unique” cell lines) displayed glucose-6-phosphate dehydrogenase (G6PD) and phosphoglucomutase (PGM) isoforms that were identical to HeLa cells (4). HeLa, established in 1951 from the cervical adenocarcinoma biopsy of a 31 year old African American woman, was the first human cell line to be successfully created in vitro (5). HeLa cells were shared with researchers all across the world and today, more than 60 years after initial propagation in culture, they continue to be a frequently used cell line for various research purposes.
HeLa is known to be an extremely aggressive and fast growing cancer cell line, and because the cell line was found in many cell culture labs from early in the 1950s, it is logical to assume that HeLa was present in many laboratories where researchers were trying to establish their own cell lines, such as Chang Liver. In fact, Dr Chang published several papers utilizing HeLa cells during the 1950s, confirming its presence in his lab during the time that Chang Liver was allegedly established (6-8).
Chang Liver: Chromosomal Analysis Supports Evidence of HeLa Contamination
In 1974, in the wake of the controversy, Dr Chang submitted a culture of “Chang Liver” cells to Dr Walter Nelson-Rees, at the University of California, for chromosome analysis (9). Nelson-Rees found that the “Chang Liver” cell line exhibited a group of distinctive chromosomes which were specific to the HeLa cell line (10).
A 1976 publication by Lavappa et al confirmed Nelson-Rees’ chromosomal observations. Tests were performed by staff at the ATCC, using Dr Chang’s deposit (11). In 1977, O’Brien et al. published information that included the genotype frequency of HeLa as 0.013 (based on the allelic frequencies of the 7 enzyme loci tested); the probability that another cell line would express the same genotype was calculated as 0.05 to 1. They found “Chang Liver” cells, received from the ATCC, had identical gene-enzyme systems when compared to HeLa and three other previously identified HeLa-contaminated cell lines (HEp-2, KB, and J111; Ref. 12).
Chang Liver: Proof of liver origin or just dedifferentiation?
In an August 1977 letter to Science (3), Dr Chang argues that “Chang Liver” expresses liver-like properties, as reported by several other researchers:
- 1971- Kaighn et al found that “Chang Liver” was able to “synthesize and secrete an antigen identical to human serum albumin…” and that “cell lines from nonliver sources do not exhibit this property”, a finding that was unexpected even to the authors due to the controversy over the authenticity of “Chang Liver” (13).
- 1974 – Bausher et al found that “Chang Liver” exhibited tyrosine aminotransferase activity, an enzyme present in the liver (14).
- 1977 – Ludueña et al presented evidence that “Chang Liver” synthesized liver alkaline phosphatase, but HeLa cells, tested in tandem, did not (15).
In response to Dr Chang’s argument, Dr Nelson-Rees commented on several publications that support unexpected phenotypic findings in cell lines known to be HeLa contaminated (16):
- 1977 – Nelson-Rees’ own correspondence in Cancer Research explains that two cell lines, HT-39 and G-11, both HeLa contaminated cell lines originally thought to be derived from carcinomas of the breast, produced α-lactalbumin, a product that, in vivo, is made by the mammary duct cells (17).
- 1977 – Ofner et al. showed that MA160, another HeLa contaminated cell line originally thought to be prostate in origin, displayed C19-steroid-metabolizing enzymes and pathways found in the human prostate (18).
The explanation for these unusual findings, surmised by Nelson-Rees, was that HeLa cells have the potential to produce such phenotypic alterations following “long-term cultivation” and “depending on the particular strain used and the stimuli applied” (17).
Today, the term dedifferentiation explains a phenomenon where tumor cells regress from a specialized function to a simpler state reminiscent of stem cells (19). Van Staveren et al studied a panel of thyroid tumor cell lines and found gene expression profiles closely resembling each other rather than the in vivo differentiated tumors from which they were derived. These cell lines exhibited phenotypic characteristics of fully dedifferentiated carcinomas, having lost expression of their thyroid-specific genes, in part, due to the absence of thyroid-stimulating hormone (thyrotropin; TSH) in the culture media (20).
Dedifferentiation may be a logical explanation for the phenotypic changes reported for the different HeLa contaminated strains.
Chang Liver: Still used today as an in vitro liver model
Advances in human DNA identification, namely the use of short tandem repeats (STRs; DNA fingerprinting), for the purpose of human cell line authentication, have made it possible to definitively address cross-contamination controversies, such as the one presented here. In a 2001 publication by Masters et al, the STR profile of “Chang Liver” was found to definitively match the HeLa cell line, making this a controversy no more (21).
ATCC continues to sell “Chang Liver” under its original catalogue number (now more appropriately termed “HeLa [Chang Liver]”); the description includes the warning that it is, in fact, a HeLa derivative:
“This line was originally thought to be derived from normal liver tissue, but was subsequently found, based on isoenzyme analysis, HeLa marker chromosomes, and DNA fingerprinting, to have been established via HeLa cell contamination”
(http://www.atcc.org/products/all/CCL-13.aspx#characteristics).
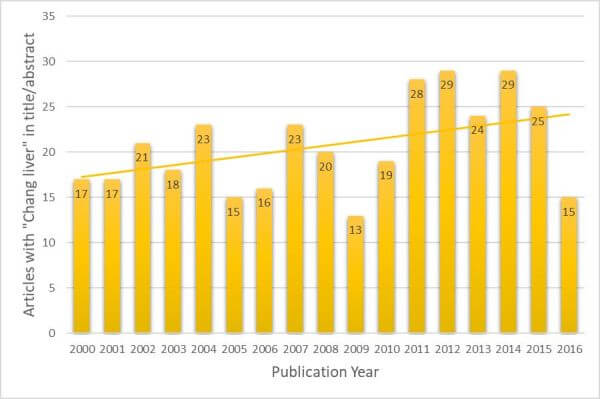
A PubMed search was performed, looking for articles with “Chang Liver” in the title or abstract (excluding the use of “Chang” or “liver” that are separate to this cell line). It was found that from the year 2000 through the end of 2016, there were 352 articles published which inappropriately used “Chang Liver” as a liver model; only one abstract, in this same time frame, stated that Chang Liver was in fact HeLa. The trend of papers published per year has not gone down but, in fact, seems to be slightly increasing (Figure 1). This is consistent with findings from other authors (22).
Figure 1. Articles that refer to “Chang liver” cells in Title or Abstract by Publication Year.
This dataset was assembled by searching PubMed for “Chang” and “liver (Title/Abstract only).
The search result was curated by removing articles that did not refer to the cell line.
Summary
Dr Chang, in that 1977 letter to Science (3), said “to indict a cell line as a HeLa cell contaminant on insufficient evidence may be counterproductive.” We now have several decades’ worth of solid evidence, including a definitive STR DNA profile match, which supports the identity of “Chang Liver” as misidentified HeLa cells.
The continued use of “Chang Liver” as a model of normal liver is what is counterproductive to the goal of advancing our understanding of cancer and the basic mechanisms that drive it.
References
1. Chang RS et al. (1971) J Natl Cancer Inst 47(2):479-83.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/4326938
2. Chang RS (1954) Proc Soc Exp Biol Med 87(2):440-3.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/13237268
3. Chang RS (1978) Science 199(4328):567-8.
Journal website | http://science.sciencemag.org/content/199/4328/567.2
4. Gartler SM (1967) Natl Cancer Inst Monogr 26:167-95.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/4864103
5. Gey GO et al. (1952) Cancer Research 12(4): 264-265.
6. Geyer RP, Chang RS (1957) Proc Soc Exp Biol Med 95(2):315-8.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/13441722
7. Chang RS (1957) Proc Soc Exp Biol Med 96(3):818-20.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/13505869
8. Chang RS (1958) Proc Soc Exp Biol Med 99(1):99-102.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/13601780
9. Nelson-Rees WA, Flandermeyer RR (1976) Science 191(4222):96-8.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/1246601
10. Miller OJ et al. (1971) Cytogenetics 10(5):338-46.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/5156367
11. Lavappa KS et al. (1976) Nature 259(5540):211-3.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/1250349
12. O’Brien SU et al. (1977) Science 195(4284):1345-8.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/841332
13. Kaighn ME, Prince AM (1971) Proc Natl Acad Sci U S A 68(10):2396-400.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/5002818
14. Bausher J, Schaeffer WI (1974) In Vitro 9(4):286-93.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/4157201
15. Ludueña MA et al. (1977) J Cell Physiol 91(1):119-29.
Journal website | http://onlinelibrary.wiley.com/doi/10.1002/jcp.1040910112/abstract
16. Nelson-Rees WA in reply to Chang RS (1978) Science 199(4328):567-8.
Journal website | http://science.sciencemag.org/content/199/4328/567.2
17. Nelson-Rees WA (1977) Cancer Res 37(9):3464-5.
Journal website | http://cancerres.aacrjournals.org/content/37/9/3464.full.pdf
18. Ofner P et al. (1977) In Vitro 13(6):378-88.
Journal website | http://rd.springer.com/article/10.1007%2FBF02615098
19. Jögi A et al. (2012) Ups J Med Sci 117(2):217-24.
PubMed Central | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3339553/
20. van Staveren WC et al. (2007) Cancer Res 67(17):8113-20.
PubMed | http://www.ncbi.nlm.nih.gov/pubmed/17804723
21. Masters JR et al. (2001) Proc Natl Acad Sci U S A 98(14):8012-7.
Journal website | http://www.pnas.org/content/98/14/8012.full
22. Gao Q et al. (2011) Hepatology 54(5):1894-5.
PubMed | https://www.ncbi.nlm.nih.gov/pubmed/21656536